
WSRC-MS-2000-00306
Biologä ID as
Compared to 16S Ribosomal RNA ID for
Environmental Isolates from the Deep Subsurface
P. C. McKinsey, M. M. Franck, and C. B. Fliermans
Westinghouse Savannah River Company
Aiken, SC 29808
D. J. Addis
Medical College of Georgia
Augusta, GA
K. B. Maxwell
Oak Ridge Institute for Science and Engineering
Oak Ridge, TN
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
This report has been reproduced directly from the best available copy.
Available for sale to the public, in paper, from: U.S. Department of Commerce, National Technical Information Service, 5285 Port Royal Road, Springfield, VA 22161, phone: (800) 553-6847, fax: (703) 605-6900, email: orders@ntis.fedworld.gov online ordering: http://www.ntis.gov/support/ordering.htm
Available electronically at http://www.osti.gov/bridge/
Available for a processing fee to U.S. Department of Energy and its contractors, in paper, from: U.S. Department of Energy, Office of Scientific and Technical Information, P.O. Box 62, Oak Ridge, TN 37831-0062, phone: (865 ) 576-8401, fax: (865) 576-5728, email: reports@adonis.osti.gov
Abstract
The U.S. Dept of Energy (DOE) Subsurface Microbial Culture Collection (SMCC) contains nearly 10,000 strains of microorganisms isolated from terrestrial subsurface environments. Many of the aerobic, Gram-negative, chemoheterotrophs isolated from the DOE Savannah River Site (SRS) have previously been identified by phylogenetic analysis of 16S ribosomal RNA (rRNA) gene nucleotide sequences. These SMCC isolates are currently being examined using Biolog GN Microplatesä and the Biolog Microstation Systemä in order to gain knowledge of their metabolic capabilities and to compare Biologä IDs with 16S IDs. To accommodate the particular needs of these subsurface isolates, which are often incapable of growing under high-nutrient conditions, Biolog’s recommendations for inoculating isolates into Biolog GN Microplatesä have been altered. The isolates are grown on low nutrient media, sodium thioglycolate (3mM) is added to the culture media to inhibit capsule formation, and a low density of bacteria is inoculated into the microplate. Using these altered inoculation criteria, 60% of these SMCC isolates have a Biologä genus ID that matches the 16S rRNA ID. These results indicate that the Biolog Systemä can be a good means of identifying unusual environmental isolates, even when recommended inoculation procedures are altered to accommodate particular isolate needs.
Introduction
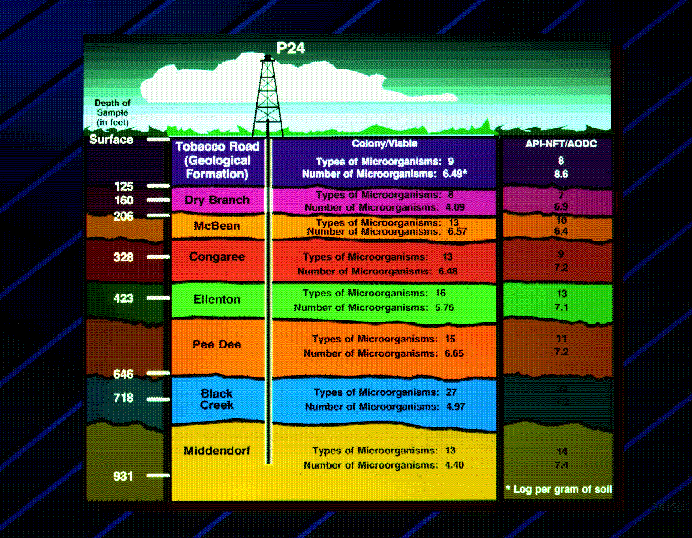
The U.S. Dept of Energy (DOE) Subsurface Microbial Culture Collection (SMCC) contains nearly 10,000 strains of microorganisms isolated from terrestrial subsurface environments. Many of the cultures in the collection were isolated during subsurface drilling at the Savannah River Site (SRS) near Aiken, SC. During drilling, sterilized plexiglass insert tubes sealed with Teflon were placed into the sampling barrels and tracer technologies were incorporated to insure the microbiological integrity of the cored sediments. Sediments were removed from the sampling barrel and immediately extruded into a nitrogen-flushed glove bag. The outer two-thirds of each core was aseptically pared away, and only the core’s center portion was used for microbiological study (Phelps et al. 1987, 1989, Fliermans and Balkwill, 1989).
The SMCC isolates examined in this study are aerobic, Gram-negative, chemoheterotrophs. They were isolated from samples collected in Savannah River Site borehole P24 at depths ranging from 91 meters to 259 meters. The SMCC isolates studied were previously characterized by phylogenetic analysis of 16S ribosomal RNA gene nucleotide sequences by Balkwill et al. (1997) using 16S rDNA as the template for PCR amplification of the 16S rRNA gene. The Gram-negative isolates studied have a 16S rRNA genus ID in one of five genera – Comamonas, Acinetobacter, Pseudomonas, Alcaligenes, and Sphingomonas. Since these subsurface isolates are often incapable of growing in high-nutrient conditions, they are routinely grown in our laboratory on lower nutrient media - 1% PTYG (0.10 grams glucose, 0.10 grams yeast extract, 0.05 grams peptone, 0.05 grams tryptone, 0.6 grams magnesium sulfate, 0.07 grams calcium chloride, 15 grams Bacto granulated agar per liter) or 50% TSA (7.5 grams tryptone, 2.5 grams soytone, 2.5 grams sodium chloride, 15 grams Bacto granulated agar per liter)
Materials and Methods
Before inoculation into Biolog GN Microplatesä , SMCC isolates were grown at 25oC on 50% TSA with sterile 3mM sodium thioglycolate swabbed onto the plate surface. Sodium thioglycolate was used to inhibit capsule formation since these isolates often form capsules when grown on laboratory media. The capsular material frequently caused false positives wells when capsule formation was not controlled in the inoculum. When sufficient plate growth was noted (SMCC isolates are often slow growers and can take up to a week for abundant plate growth), the isolates were suspended in 0.85% saline and inoculated into Biolog GN Microplatesä (150ul per well) at a transmittance level of 70% (+/- 4%). More turbid inoculum routinely caused false positive wells over the entire plate (including A1). The Biolog plates were incubated in Whirlpack bags at 25oC for varying incubation times, depending upon the incubation time required for sufficient growth of the isolate on 50% TSA. Biologä plates were read using the Biolog Microstation Systemä and Biolog ML3ä software. Biolog’s identification and similarity coefficient were noted, and the Biologä genus ID was compared to the 16s genus ID. If Biologä identified more than one species per genus, the similarity coefficients of the species were added to give a genus-only similarity coefficient.
Results
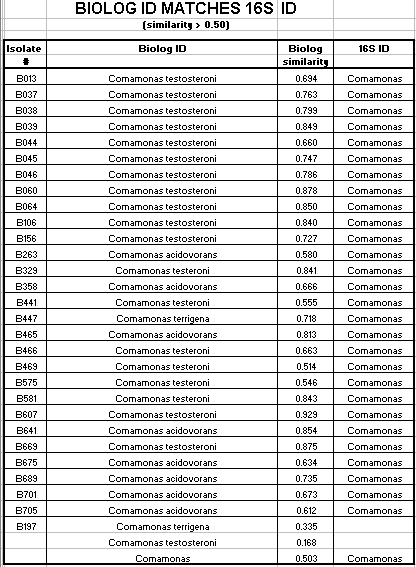
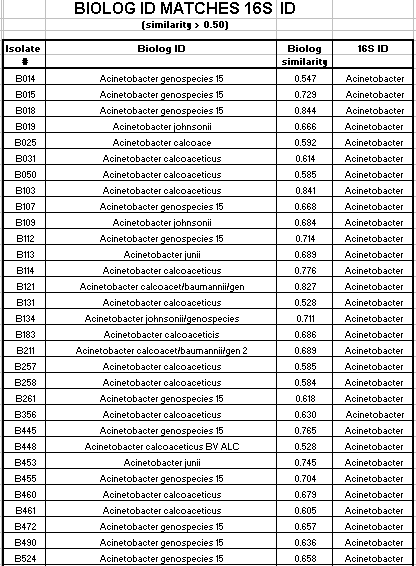
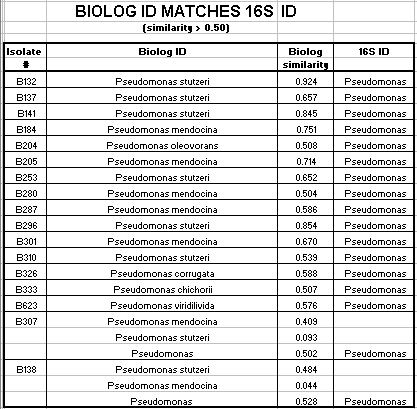
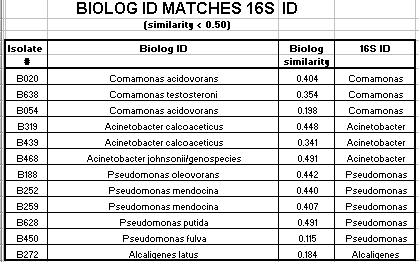
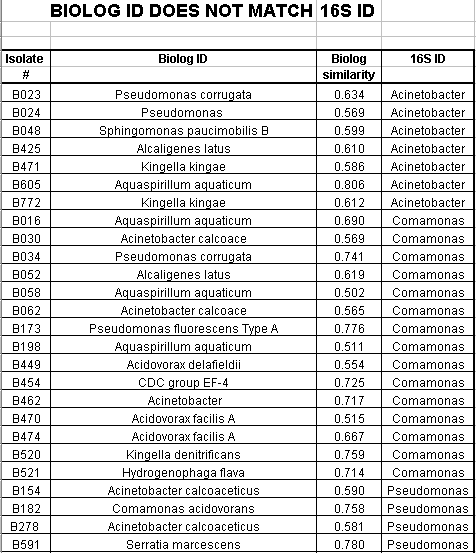
Of the 124 isolates tested, 82 isolates had a Biologä ID that corresponded to the 16s rRNA ID. These 82 isolates had a Biologä similarity value of greater than 50%. 12 additional isolates had a Biologä ID that also corresponded to the 16s rRNA ID, but the Biologä similarity value was less than 50%. Biologä does not consider identification conclusive if the Biologä similarity value is less than 50%. The Biologä ID did not correspond to the 16S rRNA ID for 30 of the 124 isolates tested.
Of the Comamonas isolates tested, 29 of 47 (62%) had a matching 16s rRNA ID and Biologä ID with a Biologä similarity value of greater than 50%. Of the Acinetobacter isolates tested, 36 of 46 (78%) had a matching16s rRNA ID and Biologä ID with a Biologä similarity value of greater than 50%. Of the Pseudomonas isolates tested, 17 of 27 (63%) had a matching16s rRNA ID and Biologä ID with a Biologä similarity value of greater than 50%. Only 2 of the 124 isolates had an Alcaligenes 16s rRNA ID. Neither isolate had a matching Biologä ID with a similarity over 50%. Also only 2 of the 124 isolates had a Sphingomonas 16s rRNA ID. Neither of these isolates had a matching Biologä ID.
In summary, 66% of the SMCC isolates had a matching 16s rRNA ID and Biologä ID with a similarity value of greater than 50%. Additionally, 10% of the isolates had a matching16s rRNA ID and Biologä ID with a similarity value of less than 50%. 24% of the isolates did not have a matching 16s rRNA ID and Biologä ID.
Conclusions
When testing SMCC subsurface isolates with the Biologä system, the intent was to examine metabolic capabilities, potentials, and similarities. Since these subsurface isolates have unique growth requirements and often produce capsules that interfere with the Biologä pattern, a unique "SMCC inoculum protocol" was created which minimized false positive plates and produced reproducible plate patterns. In order to obtain accurate identifications of bacterial strains, Biologä recommends closely following their inoculum recommendations which include growth of GN isolates on TSA with 5% sheep blood and an inoculum transmittance level of 53% to 59%. Application of sodium thioglycolate to the plate surface is not a routine Biologä recommended procedure. Because of the significant differences in Biologä recommended inoculum preparation and our "SMCC inoculum protocol", a high similarity between the Biologä genus ID and the 16S genus ID was not expected. Furthermore, since these unique subsurface environmental isolates have never previously been characterized by the Biologä system, it seemed unlikely that the Biologä system could correctly identify these unique bacterial types.
Yet when comparing the Biologä genus ID and the 16S genus ID, a greater than 60% similarity between the two identifications was noted. Of the isolates that did not have matching IDs, some had similar identifications (Comomonas/Pseudomonas) which, depending upon the species, could be synonym identifications. Biologä identified five isolates as species of Kingella, which show very few positive wells on the Biologä plate. The low number of positive wells for these particular oligotrophic isolates may be an indication of their failure to adjust to the higher concentrations of nutrients in the Biolog plates. The Biologä system would be incapable of correctly identifying these particular SMCC isolates if the Biolog reaction was not a true indication of their metabolic capabilities.
The results of this study indicate that the Biolog Systemä can be a good means of identifying unusual environmental isolates, even when recommended inoculation procedures are altered to accommodate particular isolate needs. The Biologä system is a particularly useful environmental tool since it, unlike 16S identification protocols, can identify isolates to the species level and, at the same time, can show metabolic capabilities of the isolates.
References