WSRC-MS-2000-00220
Characterization of a Kineococcus-like
Isolate
from a Radioactive Work Area
C. J. Berry and C. Fliermans
Westinghouse Savannah River Company
Aiken, SC 29801
R. W. Phillips, J. Wiegel, and L. J. Shimkets
University of Georgia
Athens, GA 30602
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
This report has been reproduced directly from the best available copy.
Available for sale to the public, in paper, from: U.S. Department of Commerce, National Technical Information Service, 5285 Port Royal Road, Springfield, VA 22161, phone: (800) 553-6847, fax: (703) 605-6900, email: orders@ntis.fedworld.gov online ordering: http://www.ntis.gov/support/ordering.htm
Available electronically at http://www.osti.gov/bridge/
Available for a processing fee to U.S. Department
of Energy and its contractors, in paper, from: U.S. Department of Energy, Office
of Scientific and Technical Information, P.O. Box 62, Oak Ridge, TN 37831-0062,
phone: (865 ) 576-8401, fax: (865) 576-5728, email: reports@adonis.osti.gov
Abstract
A Kineococcus-like bacterial isolate designated SRS30216 has been recovered from a radioactive work area at Savannah River Site in Aiken, S.C. The sequence of the 16S rRNA gene from SRS30216 is 93% identical to that of Kineococcus auranticaus RA333T. SRS30216 and K. aurantiacus RA333T display an intermediate level of resistance to gamma-irradiation and desiccation compared to Deinococcus radiodurans R1.
The search for bacteria in novel environments has led to the discovery of organisms that can live under conditions of extreme temperatures, pH levels, salinity, and pressures (5, 6, 11). New organisms continue to be isolated from these novel environments, adding to the fast growing number of prokaryotic species. We have investigated the possibility that bacteria grow in a radioactive environment at the Savannah River Site (SRS) in Aiken, South Carolina. At SRS, high level radioactive waste is stored in basins where the gamma radiation dose can be as high as 0.1 kGray/hour (1 kGray = 105 rads). The possibility that organisms grow under these conditions is thought to be unlikely. However, organisms such as Deinococcus radiodurans R1 (1) and Rubrobacter radiotolerans (14), which can withstand short exposures to extreme doses of gamma radiation, suggests the existence of bacteria that can survive constant exposure to gamma radiation. A bacterial isolate designated SRS30216 was obtained using a sterile swab to sample the floor of a high level radiation work area at SRS. The radiation levels in the work area fluctuate within a range of 18 rads/hour to greater than 350 rads/hour. Based on 16S rDNA sequence, SRS30216 is related to microorganisms in the genus Kineococcus (13). The only member of this recently described genus of gram type-positive microorganisms is K. auranticus RA333T (13). K. auranticus is a motile, coccus-shaped organism, isolated from soil from the Indore region of India. Based on the 16S rDNA sequence, microorganisms in the genus Kineococcus are phylogenetically related to the actinobacteria but form a unique branch with few close relatives (9, 13). Although some biochemical studies have been performed with RA333T (3, 4, 13), little is know about the phenotypic properties of this genus.
Studies were carried out to characterize and compare the morphology and radiation and desiccation resistance of SRS30216 to the Kineococcus aurantiacus type strain RA333T as well as the known radiation resistant and radiation sensitive organisms Deinococcus radiodurans R1 and Escherichia coli, respectively. All bacteria were maintained in PTYG broth (10g glucose, 5g yeast extract, 5g tryptone, 5g peptone, 0.6g MgSO4•7H20, 0.06g CaCl2 per liter, 15g of Bacto agar was added to solidify media for PTYG plates). Deinococcus radiodurans R1 (ATCC 13939) and Kineococcus auranticus RA333T (ATCC 51238) were obtained from the American Type Culture Collection. Escherichia coli CF1648 (recA+) was obtained from M. Cashel. All cultures were grown on plates at 32° C or in broth cultures at 32° C with aeration.
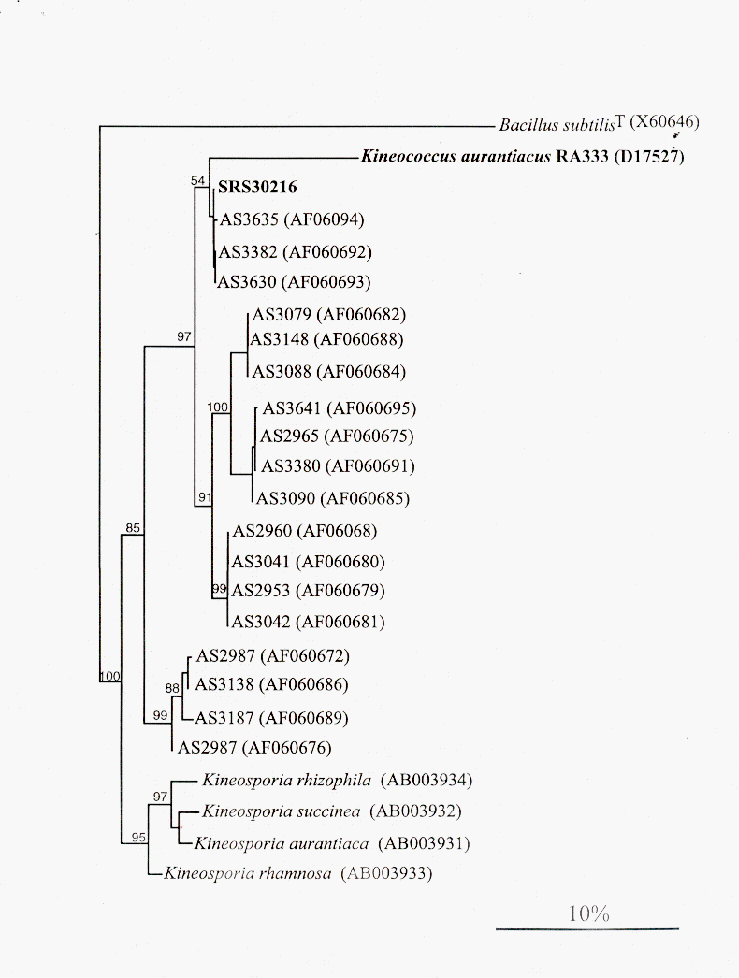
To help identify SRS30216, genomic DNA was isolated utilizing the CTAB/NaCl isolation procedure (8). The 16S rRNA gene of SRS30216 was amplified using the universal primers 27F (5’AGAGTTTGATCMTGGCTCAG3’;M=C:A) and 1392R (5’ACGGGCGGTGTGTRC3’;R=A:G) for the eubacteria 16S rRNA gene as previously described (12). Both strands of the PCR product were sequenced (Molecular Genetics Instrumentation Facility, UGA). The 16S rDNA sequence for SRS30216 shows 93% identity with the Kineococcus auranticus type strain RA333T over 1268 base pairs internal to the 16S rRNA gene. The SRS30216 16S rDNA sequence also contains a duplication of 49 nucleotides corresponding to bases 736-785 of the Escherichia coli 16S rRNA gene. Phylogenetic analysis was performed comparing the 16S gene sequence of SRS30216, RA333T and newly deposited sequences from Kineococcus-like organisms isolated from the Mojave Desert using the DNA parsimony (Figure 1) and DNA maximum-likelihood programs (data not shown) within the PHYLIP phylogeny package (2). Both algorithms show that SRS30216 and some of the Kineococcus-like organisms isolated from the desert are more closely related to each other than to the K. auranticacus type strain RA333T.
RA333T has previously been described as growing in tetrads or larger clumps of cells. Phase contrast microscopy of broth cultures suggested the same was true of SRS30216. Scanning electron microscopy was used to look at the morphology of SRS30216 after growth on PTYG medium for 3 days. Cells were scraped from the plate and washed once in phosphate buffer (4.73g Na2HPO4, 4.5g KH2PO4 per liter, pH 7.0, 0.067M final) and resuspended in 100 microliters of the same buffer. An equal volume of glutaraldehyde was added to the cells for 1 hour. The cells were then washed 3x with phosphate buffer and collected on millipore filters before being serially dehydrated with ethanol using 20% steps ending in 3 changes at 100%. Critical point drying of the samples was performed in a Samdri CPD before observation by a scanning electron microscope (LEO 982 Field emission SEM). Large packets of cells were observed with a morphology similar to that described by Yakota et al. (13). The packets appear to be made up of coccoid shaped cells that have divided in two planes (Figure 2).
Because SRS30216 was isolated from a radioactive work area, the radiation resistance of this strain was compared to the K. aurantiacus RA333T and the known radiation resistant organism Deinococcus radiodurans R1. Exponentially growing cultures were washed and resuspended in an equal volume of 0.067M phosphate buffer and divided into 100 microliter aliquots. Cells were exposed to a Co60 source for predetermined times. At each time point, 3 individual aliquots of each strain were removed from the radiation source. Cell suspensions were serial diluted in phosphate buffer and plated on PTYG medium. After 3 days of growth, colony-forming units were counted and the percent survival was calculated based upon the number of colony forming units present before irradiation. The two Kineococcus strains, SRS30216 and RA333T, showed an intermediate amount of radiation resistance compared to the radiation resistant organism D. radiodurans R1 and the radiation sensitive organism E. coli CF1648 (Figure 3).
Organisms that are radiation resistant have also been shown to be resistant to desiccation, a characteristic which may actually be more physiologically relevant (7, 10). Because SRS30216 is related to the soil organism RA333T and the Kineococcus-like desert isolates, desiccation resistance was examined. Exponentially growing cells were washed once and resuspended in an equal volume of phosphate buffer. Desiccation resistance was tested by drying 1 milliliter aliquots of D. radiodurans R1, E. coli CF1648, SRS30216 and K. auranticus RA333T onto glass cover slips (1 inch x 1 inch). The cover slips were then placed in sterile petri dishes inside a desiccator containing calcium sulfate under a vacuum. An electronic hygrometer (Fisher scientific, Cat. #11-661-14) measured the humidity at 7% +/- 2%. The percent survival for each strain was determined at 3, 7 and 14 days after desiccation. At each time point, one cover slip containing each strain was removed from the desiccator. One milliliter of phosphate buffer was added to the cover slips to rehydrate the cells. 10-fold serial dilution and plating was then used to determine the percent survival. Over a two-week period the survival of all four strains decreased with D. radiodurans R1 showing the most resistance and E. coli CF1648 showing the least resistance to desiccation. The two Kineococcus strains SRS30216 and RA333T showed an intermediate level of resistance with a level of survival of approximately one log lower than that shown by D. radiodurans (Figure 4).
In summary, we have isolated a bacterium from a high level radiation work area at the Savannah River Site in Aiken, South Carolina. Based on 16S rDNA sequence analysis, SRS30216 is phylogenetically related to Kineococcus aurantiacus RA333T. SRS30216 and the Kineococcus aurantiacus type strain RA333 show an intermediate level of radiation resistance compared to Deinococcus radiodurans R1 and E. coli. The level of resistance shown is consistent however with the lower intensity of gamma-radiation present in the environment from which SRS30216 was isolated compared to the Co60 source used in this study. Both SRS30216 and RA333T show a much higher level of resistance to desiccation than that shown by E. coli and only slightly less than that shown by D. radiodurans R1. The desiccation resistance of SRS30216 should not be surprising based on the knowledge that SRS30216 was isolated from a work room floor and the phylogenetically related Kineococcus-like strains were isolated from the Mojave desert. Furthermore, these data provide further evidence for the proposed relationship between desiccation resistance and resistance to gamma radiation which is the ability to repair damaged DNA through a recA dependent process. If desiccation resistance is a natural phenotype for these organisms, then the ability to survive desiccation may have allowed SRS30216 to survive in the presence of the high levels of radiation at SRS.
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant No. DEB 9817825.
References

Figure 1. Phylogenetic analysis of the 16S rDNA sequences from SRS30216, K. aurantiacus RA333T and selected Kineococcus-like desert isolates using the DNA parsimony algorithm. Analysis using the DNA maximum-likelihood method produced a nearly identical tree. Bootstrap values are given for major divisions. Accession numbers are in parentheses.
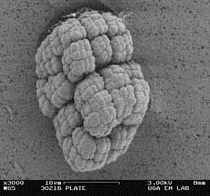
Figure 2. Scanning electron micrograph of SRS30216 cultured
on PTYG agar
showing the clusters formed by growing cells. Bar = 10 microns
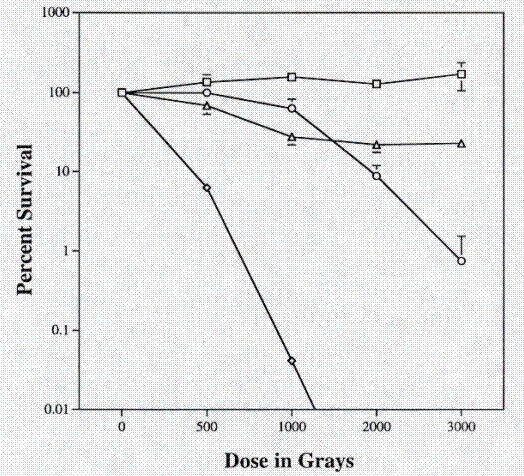
Figure 3. Resistance to gamma radiation from a Co60 source. Open square, Deinococcus radiodurans R1; closed diamond, Escherichia coli CF1648; open circle, Kineococcus aurantiacus RA333T; closed triangle, SRS30216.
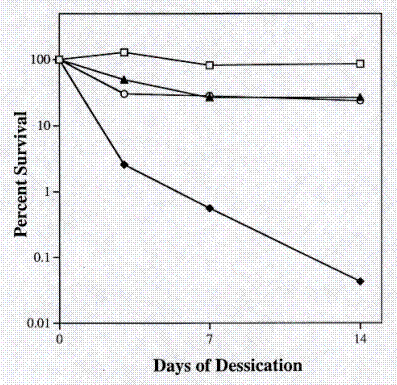
Figure 4. Resistance to desiccation. Open square, Deinococcus radiodurans R1; closed diamond, Escherichia coli CF1648; open circle, Kineococcus aurantiacus RA333T; closed triangle, SRS30216.