WSRC-MS-2001-00058
Methanotrophic Bacteria: Use in Bioremediation
R. L. Brigmon
Westinghouse Savannah River Company
Aiken, SC 29808
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
This report has been reproduced directly from the best available copy.
Available for sale to the public, in paper, from: U.S. Department of Commerce, National Technical Information Service, 5285 Port Royal Road, Springfield, VA 22161, phone: (800) 553-6847, fax: (703) 605-6900, email: orders@ntis.fedworld.gov online ordering: http://www.ntis.gov/support/ordering.htm
Available electronically at http://www.osti.gov/bridge/
Available for a processing fee to U.S. Department of Energy and its contractors, in paper, from: U.S. Department of Energy, Office of Scientific and Technical Information, P.O. Box 62, Oak Ridge, TN 37831-0062, phone: (865 ) 576-8401, fax: (865) 576-5728, email: reports@adonis.osti.gov
Glossary
The methanotrophs are aerobic bacteria that oxidize methane as an energy source and carbon source through the enzyme methane monooxygenase (MMO). This MMO can cometabolize or transform nongrowth substrates by either growing or resting cells. Cometabolism is a result of nonspecific MMO activity towards organic compounds that do not serve as carbon or energy sources. While many cometabolizing bacterial species have been identified, the best studied are the methanotrophs. The reason for this is that methanotrophs are ubiquitous and can cometabolize many aliphatic compounds, alkanes, and aromatic compounds. Methanotrophs have been intensely studied for use in degrading chlorinated solvents, most notably trichloroethylene, to environmentally acceptable concentrations in soils, sediment, and groundwater. Stimulation of methanotrophic bacteria is accomplished through the addition of methane and other gaseous nutrients resulting in an increase in contaminant biodegradation and biotransformation. The composition of gaseous nutrients used with methane is dependent on the characteristics of the site geochemistry and microbiology. This biostimulation may be applied in situ within the contaminated aquifer or soil. If necessary, the contaminated soil or groundwater can be moved and treated ex situ based on the site-specific needs.
I. Introduction
Some types of anaerobic microorganisms produce methane in environments where oxygen is limited that include swamps, landfills, wetlands, peat bogs, sediments, and the intestinal flora of animals. Methane oxidizing bacteria (methanotrophs) in turn remove methane from these environments. Therefore anaerobic sediments are considered an aerobic sink for methane due to the activities of methanotrophs.
Methanotrophs are a unique group of methylotrophic bacteria, which utilize methane as their sole carbon and energy source (Hanson and Hanson 1996, Murrell 1994). These organisms have been isolated from a wide variety of environments including soils (Whittenbury et al, 1970), sediments (Smith, et al., 1997), landfills (Wise et al., 2000), groundwater (Fliermans et al., 1988), seawater (Holmes et al., 1995, and Murrell and Holmes 1995, Sieburth et al., 1997), peat bogs (Dedysh, et al., 1998, McDonald, et al., 1996 and Ritchie, et al, 1997), hotsprings (Bodrossy, et al. 1995 and 1997), plant rhizosphere (Gilber et al., 1998), salt reservoirs (Khmelenina et al, 1996) and the Antarctic (Bowman et al., 1997).
Methanotrophs were initially grouped according to their morphology, type of resting stage, intracytoplasmic membrane structure and physiological characteristics (Whittenbury et al, 1970). Subsequent 16S rDNA sequence analysis has further clarified these phylogenetic relationships and defined eight genera of methanotrophs, namely Methylococcus. Methylomonas, Methylomicrobium, Methylobacter, Methylocaldum, Methylosphaera, Methylocystis and Methylosinus. These genera are divided into two distinct physiological groups. Type I methanotrophs (Methylomonas, Methylomicrobium, Methylobacter, Methylocaldum, Methylosphaera) assimilate formaldehyde produced from the oxidation of methane (via methanol) using the ribulose monophosphate pathway, have cellular membranes that are composed of predominantly 16-carbon fatty acids and possesses bundles of intracytoplasmic membranes. Type II methanotrophs (Methylocystis and Methylosinus) utilize the serine pathway for formaldehyde assimilation, have intracytoplasmic membranes arranged around the periphery of the cell and contain predominantly 18-carbon fatty acids (Hanson and Hanson 1996). Membranes of the genus Methylococcus posses a combination of characteristics of both type I and type II methanotrophs.
Methanotrophs form coherent phylogenetic clusters that share the common physiological characteristics described above (Murrell et al. 1998). Type I methanotrophs (including Methylococcus) cluster in the g -subdivision of the class Proteobacteria, while type II methanotrophs are grouped within the a -subdivision of the Proteobacteria. The tight phylogenetic clustering of these groups has allowed the design of a range of oligonucleotides, which target a broad range of both methanotrophs and methylotrophs (Brusseau, et al., 1994, Hanson, et al., 1993, Tsien, et al., 1990). A number of 16S ribosomal and gene functional probes are now available for culture-independent detection of methanotrophs and methylotrophs (Wise et al. 2000).
Several species of methanotrophs have been isolated that produce methane monooxygenase and participate in the cometabolic degradation of TCE (Koh et al., 1993). However, soluble methane monooxygenase (sMMO) is present in some methanotrophic bacteria that has been found to be responsible for oxidizing a wide range of carbon substrates (Cardy et al., 1991). No sMMO has been isolated or observed in type I methanotrophs (Soh et al., 1993). Moreover it appears that the sMMO is limited to a few species of type II and type X methanotrophs. The sMMO has been found through its non-specific activity to effect oxidations, dechlorinations, condensations, and rearrangements of meta-chlorotoluene, phenol, chlorofluorobenzenes, and mono and dichlorobiphenyls via itís non-specific activity. Soluble MMO is believed to be found only in type II and type X methanotrophs during copper limiting conditions (Murrell, 1992). Although membrane and particle associated MMO has been reported, the rates of chlorinated solvent degradation with these organisms is significantly less than sMMO bacteria (DiSpirito et al., 1992). These solvents have included halogenated aliphatic compounds such as trichloroethylene (TCE). Methanotrophs can utilize nitrogen as nitrate, ammonia, and molecular nitrogen while demonstrating TCE transformation capacity (Chu and Alvarez Cohen 1996). This article will concentrate primarily on methanotrophic bacteria applications shown to be effective in chlorinated solvent bioremediation with a focus on TCE.
II. Background
In recent years there has been an increased interest in the use of microorganisms for environmental restoration. The usefulness of microorganisms with a diversity of metabolic activities in wide ranging applications coupled with advances in the technology have lead to successful demonstrations in the ever-expanding bioremediation field. The application of environmental biotechnology as a successful remediation tool depends on the ability to stimulate or enhance specific activity of indigenous or introduced microorganisms. The challenge has been to enhance the activity of these microorganisms and develop means to bring the contaminant into direct contact with the organisms to achieve optimal bioremediation. Methanotrophic bacteria have a ubiquitous distribution in the environment and the use of natural gas or methane with other nutrients to stimulate their bioremediation activities through methane monooxygenase is a remediation option. These two features allow for a relatively efficient, inexpensive, and safe means to manipulate the environment to accelerate bioremediation. This section describes the great interest in methanotrophic bacteria in bioremediation.
TCE is a volatile chlorinated organic compound that has been widely used as an organic solvent and degreasing agent and disseminates over large areas in the subsurface at contaminated sites. TCE is the most frequently observed volatile organic compound (VOC) at Resource Conservation and Recovery Act (RCRA) sites (Alvarez-Cohen and McCarty, 1991) and in groundwater (Westrick et al. 1984). TCE is a VOC that is regulated as an air pollutant under the Clean Air Act Amendments Title III. The U.S. Occupational Safety and Health Administration is setting worker exposure limits for this solvent due to itís toxicity. Removing TCE from contaminated groundwater can pose risks using conventional engineering methods such as vacuum extraction or pump and treat remediation. These risks include transfer of TCE to air through volatilization and handling issues. Due to increasing environmental concerns and federal regulations there is a growing preference for bioremediation applications that reduce risk and produce minimal toxic residuals. Although investigations of the microbial degradation of chlorinated ethenes have been ongoing since the 1950ís (Davis, 1956), as recent as the 1980ís it was believed that TCE could not be successfully biodegraded either aerobically or anaerobically (Bower et al., 1984). This belief was in part due to the wide spread groundwater contamination by TCE. In 1980, California closed 39 public water supply wells in the San Gabriel Valley because of TCE pollution, and New York, New Jersey, and Pennsylvania condemned wells because of the same contaminant (Metzler, 1982).
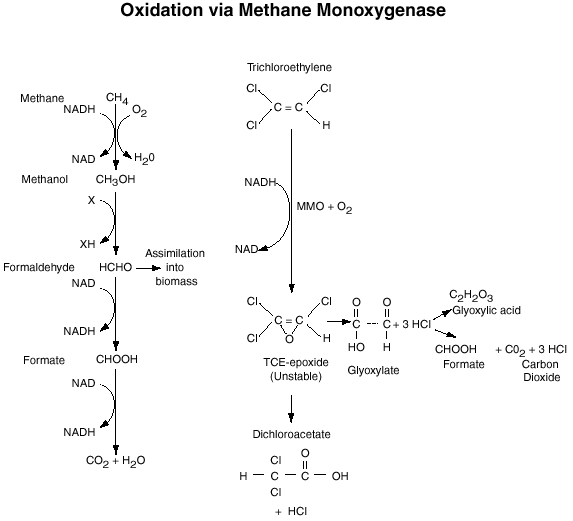
Wilson and Wilson (1985) demonstrated that TCE is susceptible to cometabolism by soil communities enriched with natural gas. Fliermans et al., (1988) and others demonstrated that cultures enriched with methane and propane could cometabolically degrade a wide variety of chlorinated aliphatic hydrocarbons including ethylene, 1,2-cis-dichloroethylene (c-DCE); 1,2-trans-dichloroethylene (t-DCE); and vinyl chloride (VC); toluene; phenol and cresol. Investigations that utilized methods to stimulate microorganisms in the subsurface vadose zone and groundwater found that both aerobic and anaerobic processes contributed in removal of TCE as revealed by detection of TCE oxidation and anaerobic transformation products, c-DCE; t-DCE; and VC (Hazen et al., 1993; Fliermans et al., 1994). These two studies proved that bioremediation provides a powerful means of restoring contaminated aquifers. McCarty and Semprini (1994) concluded from laboratory and field studies that cometabolic transformation of TCE was strongly tied to methane utilization. When methane addition were stopped TCE transformation ceased. Figure 1 outlines the oxidation of methane by methane monooxygenase and associated cometabolism of trichloroethylene.
It is now well recognized that TCE and other chlorinated aliphatic compounds can be degraded by a diversity of bacteria including methanotrophs (Little et al., 1988), selected methanogens (Bouwer and McCarty, 1984), and species of Pseudomonas (P. cepacia, P. mendocina and P. putida) capable of also degrading aromatic compounds (Nelson et al., 1988). Ensley (1991) has demonstrated a linkage between TCE degradation and aromatic metabolism in P. cepacia G4, P. mendocina and P. putida. Easign et al., (1992) reported that pure cultures of Xanthobacter spp. cometabolized TCE with the utilization of propylene as a carbon and energy source presumably using the enzyme alkene monooxygenase. Fliermans, et al (1988) and Bowman et al., (1993) have shown that enrichments for methanotrophs in subsurface samples collected from the Savannah River Site in South Carolina stimulate the microbial degradation and complete mineralization of TCE and other chlorinated aliphatic compounds both in the laboratory and in situ. Propane utilizers or propanotrophs that also exhibit non-specific oxidase activity may also be used for bioremediation of VOCs. Where mixtures of chlorinated aliphatic hydrocarbons including 1,1,1-trichloroethane, are present propane may be the stimulant of choice using air-sparging technology (Tovanabootr & Semprini, 1998). However, methanotrophs are optimal in bioremediation when TCE is the primary contaminant of concern.
An advantage of utilizing methanotrophs for bioremediation is that aerobic conditions do not appear to support the formation of undesirable metabolites, such as c-DCE, t-DCE or VC that are partially dechlorinated byproducts of anaerobic degradation of TCE. VC, a known carcinogen, is considered more hazardous than the parent compound TCE. Methane oxidizers are likely to be found in zones that fluctuate between aerobic and anaerobic conditions such as soils that periodically flood and drain (Atlas and Bartha, 1987). Enzien et al, (1994) has suggested that both anaerobic and aerobic populations may both be stimulated to biodegrade TCE in an aquifer under bulk aerobic conditions.
Although there appears to be a diversity of natural microorganisms that degrade TCE, the compound remains the most prevalent organic contaminant in the United States (Vogel and McCarty, 1985). The ubiquity of the TCE contamination suggests that either the environmental conditions do not support biodegradation or that the correct consortia of microorganisms is not present, or at sufficient cell densities in the ecosystem. Thus it is important to understand both the biological as well as the physical parameters associated with biodegradation of TCE. Despite these possible problems, the promise of bioremediation is revealed by the observation that once stimulated, mixed methane-grown communities have been found to metabolize TCE completely to harmless endproducts with a half-life of less than a day (Semprini et al., 1994)
While sediment methanotrophic bacteria can be efficient in degrading TCE from contaminated groundwater (Bowman et al, 1993), certain methanotrophs are more efficient at TCE degradation than others (Koh, et al. 1993). It has been suggested that mixed populations are more efficient in TCE degradation (Uchiyama et al., 1992). Field applications of methanotrophs for removal of TCE from the environment have proven that further studies of these bacteria are needed. Specific areas that need to be explored are associated microbial communities, population dynamics, and potential for biodegradability to decrease or eliminate the contaminant. Thus a better understanding of the microbial ecology of TCE biodegradation is clearly needed.
III. In situ Bioremediation
In situ bioremediation means that the environmental restoration of contaminated sediments are not moved from the site or groundwater is not pumped and treated at the surface. When an in situ bioremediation technology is employed the relocation and transport of materials may be avoided. This makes in situ bioremediation, where applicable, a highly attractive technology for remediation because contaminants are removed in place, not simply moved to another location or volatilized (Baker & Herson, 1991). In situ bioremediation of chlorinated solvents with methane injection is a site-specific application. This application results in decreased remediation costs, lower contamination risks, shorter restoration time and increased efficiency, as well as enhanced public and regulatory acceptability. Public relations are enhanced because much of the action occurs with minimum above ground activity and equipment. Bakst (1991) has determined bioremediation to be among the least costly technologies where its application is feasible.
The Savannah River Site (SRS) is a 320 square mile facility owned by the U.S. Department of Energy and operated by Westinghouse Savannah River Company. The SRS, located in a rural area along the Savannah River in Aiken and Barnwell counties of South Carolina, has generated nuclear materials for defense, medical, and space applications since the 1950ís. During the first 20 years of operation most of the waste generated at the SRS including millions of pounds of chlorinated solvents was handled via burning rubble pits, evaporation ponds, and waste pits resulting in soil, sediment and groundwater contamination.
The SRS has now completed several successful bioremediation demonstrations on soil and groundwater contaminated with chlorinated ethenes utilizing methanotrophic biostimulation (Altman et al., 1998, Brigmon et al. 1999, Hazen et al., 1997). The deployment of two projects at SRS designed for field demonstrations of in situ treatment of groundwater contaminated with chlorinated solvents by gaseous nutrient injection have proven the effectiveness of in situ bioremediation.
A. Demonstration 1
The first demonstration was near the 300-M Area operations area where fuel and target elements were degreased in processing. An estimated 13 million pounds of solvents were used in this processing from 1952 to 1982. While evaporation was used to reduce much of the solvents an estimated 2 million pounds was released to the M Area Settling Basin (Hazen et al., 1994). These discharges to the M Area settling basin consisted primarily of TCE, perchloroethylene (PCE), and trichloroethane (TCA).
The M area in situ bioremediation demonstration consisted of two horizontal wells for injection and extraction at the process sewer line leaking PCE and TCE (Hazen et al., 1994). Figure 2 illustrates a side view of the 160-ft deep horizontal wells in relation to the surface nutrient injection and extraction systems. Groundwater, extracted air, and sediment samples were taken before, during, and after testing of the system that ran for over one year. Subsurface gaseous nutrient injection including methane, nitrogen, and phosphorous was found to be an effective in situ bioremediation treatment for TCE-contaminated groundwater at the M Area site of SRS (Hazen et al., 1997). Gene probe analysis demonstrated an increase MMO activity corresponded with significant TCE degradation rates among a high proportion of the indigenous methanotrophic bacteria in the M Area groundwater (Bowman et al., 1993). Species-specific methanotrophic bacteria DNA probes have revealed that perturbations associated with biostimulation resulted in preferential changes in the structure and physiological status of microbial communities (Brockman et al., 1994). Direct evidence of TCE mineralization in this project was demonstrated by increased groundwater chloride concentrations that correlated with several-fold increases in the density of methanotrophic bacteria (Pfiffner et al., 1997). The rate of TCE biodegradation also correlated to growth of specific species of methanotrophs and nutrient growth factors including injected methane, phosphorous and nitrogen (Travis and Rosenberg, 1997). These increases correlated with methane groundwater concentrations and the pulsed injection regime over the year test period.
The observation that methanotrophic population increases correlated with TCE biodegradation indicated that the zone of influence of gaseous nutrient injection extended at least 60 ft from the injection well in both the horizontal and vertical directions (Brockman et al., 1995). Groundwater monitoring of both bacteria and contaminant concentrations provide information on the efficiency of this technology (Fliermans et al., 1994). Application of these methane/air mixtures demonstrated a 3 to 5 order of magnitude increase in TCE-degrading methanotrophic bacteria during the methane- and nutrient-injection (Pfiffner et al., 1997).
A subsequent remediation project was conducted in the same M-Area aquifer to determine the effect of chemical oxidation on subsurface microbiology and co-metabolic biodegradation capacity in the same aquifer after treatment with Fentonís reagent (Kastner et al., 2000). The groundwater pH declined from 5 to 2.4 immediately after the Fentonís treatment indicating release of Cl- ions from TCE/PCE. The pH subsequently rose to a range of 3.4 - 4.0 after 17 months. Limited methanotrophic bacteria growth and TCE degradation were detected in the treated zone (pH 3.37 and TCE 5 mg L-1) with methane addition. Methane addition to groundwater from the control well without Fentonís Reagent, (pH 4.9 and TCE 0.7 mg L-1), stimulated methanotrophic growth. This was indicated by methane consumption and microbial characterization with fluorescent antibody analysis, phospholipid based markers, and rDNA probes. Higher TCE concentrations in the Fentonís Reagent treated zone (16-21 mg L-1) might have inhibited TCE co-metabolism. These results also indicate that low groundwater pH resulting from the chemical oxidation process (pH 3.3 vs. 4.9) inhibited TCE biodegradation. Methanotrophic growth and TCE biodegradation may be possible as pH increases both in the treated zone and at the leading edge of plume if sediments are able to buffer groundwater pH. The treatment with Fentonís limits the potential for monitored natural attenuation to occur. Additionally, the Fenton's reagent process could be designed to operate at a higher pH (e.g., ³ 4.5) and/or lower H2O2 concentration to minimize detrimental long term biological effects, providing an optimal environment to couple advanced oxidation processes with bioremediation technologies.
B. Demonstration 2
A second demonstration of in situ bioremediation at SRS is at the Non Radioactive Waste Disposal Facility (NRWDF) formerly known as the Sanitary Landfill. The NRWDF began receiving solid waste from construction areas, offices, shops, and cafeterias in 1974. During the course of its operation, the Sanitary Landfill received numerous materials that can leach or generate hazardous compounds, e.g. paints, thinners, solvents, batteries, and rags and wipes used with organic solvents. Wastes were cataloged but not segregated within the landfill. As a result in 1988, recurring evidence of contaminants of concern (COCs) was detected in the groundwater including TCE, PCE, TCA, DCE, VC, and chlorobenzene (CB).
Initially, a treatability study to evaluate the NRWDF bioremediation potential using soil columns was employed to simulate both vadose and groundwater conditions using NRWDF sediment and groundwater (Enzien et al., 1994). The results of the treatability study proved that cometabolic methanotrophic bioremediation of the COCs was possible at the NRWDF. A subsequent in situ optimization test demonstrated that biostimulation by addition of oxygen, nutrients, and methane at two sites within the NRWDF resulted in undetectable levels of contaminants and other organics in both the groundwater and vadose zone (WSRC, 1996b). Additionally chloride concentrations in the groundwater at both sites increased significantly as methanotrpphic bacteria densities increased, reaching a maximum population in 3-4 days, contaminant levels decreased.
Figure 3 demonstrates methanotrophic bacteria labeled with direct fluorescent antibodies from NRWDF groundwater concentrated on a 0.2 m m filter (Brigmon et al, 1998). The total number of groundwater microorganisms did not change, indicating a selective stimulation of the methanotrophic population. (Brigmon et al 1998). The loss of contaminants appears to be due to cometabolic biodegradation through biostimulation since loss by volatilization was minimal. This work again clearly demonstrates that one can effectively change the subsurface bacterial population in a relatively short period of time. A larger scale (400 & 600 ft) set of horizontal wells at the NRWDF is now being tested with nutrient injection for long-term containment of chlorinated ethenes (Figure 4).
Similarly CB groundwater concentrations, another NRWDF contaminant, have significantly decreased in recent years at the NRWDF with a concomitant increase in chloride ions (Brigmon & Fliermans, 1997). Chlorobenzene can undergo microbiological dechlorination and the benzene ring can be converted to catechol, followed by ring fission or oxidation of the side chain. Geochemical data confirm that TCE and CB concentrations are decreasing at a greater rate than would be expected due to NRWDF groundwater transport or dilution.
These demonstrations represent another approach in the development of in situ bioremediation technologies. In both applications a pulsed application of methane worked best for methanotroph biostimulation and contaminant degradation. Through the use of the gaseous nutrient injection system (Figure 4) the NRDWF groundwater COC concentrations are approaching minimal detection limits (5 ppb).
IV. Monitored Natural Attenuation
Monitored Natural Attenuation (MNA) is a risk management option that relies on natural biological, chemical, and physical processes to contain the spread of contamination from a source (Major et al., 1995). Containment by MNA relies on sorption, volatilization, dilution, destruction, or biotransformation of contaminants. Comparing rates of contaminant transport to rates of MNA to other methods including groundwater transport models can quantitatively assess the efficiency of this option to prevent contaminant migration in groundwater systems. If groundwater movement is faster relative to rates of contamination removal, contaminants have the potential to reach points of contact with human or wildlife populations. Conversely, if transport rates are slow relative to removal rates, contaminant migration will be more confined and less likely to reach a point of contact. Evaluating the factors mitigating contaminant transport to predetermined points of contact can assess the efficiency of MNA. Thus, this assessment includes hydrologic (rates of ground water flow), microbiologic (rates of biodegradation), and sociopolitical (points of contact) considerations.
The MNA of TCE is associated with anaerobic dechlorination as well as cometabolism. Under specific anaerobic conditions TCE can be reductively dechlorinated to less chlorinated ethenes including DCE and VC. The presence of these compounds is evidence of TCE intrinsic bioremediation. Bouwer (1994) has reported that if the amount of cis-DCE is greater than 80% of the total DCE, which it is in the case of the NRWDF, then it is a biodegradation product of TCE. MNA of TCE is associated with the accumulation of these daughter products and increase in chloride ions (Weidmeier et al., 1996).
Other microbial processes involving methanotrophs may also be of value to bioremediation efforts. For example, denitrification rates in anaerobic environments have been found to be directly dependent on the methanotrophic activity and the percent of soluble carbon produced during methane oxidation. Any of the three intermediates (methanol, formaldehyde or formate) from the methane oxidation (Figure 1) could be used as electron donors by anaerobes (Houbron et al., Wat Sci Tech, 1999, 40: 115-122).
A number of MNA investigations involving chlorinated ethene degradation have demonstrated that while anaerobic dechlorination is occurring, the anaerobic processes alone cannot fully account for the observed contaminant reduction. Sequential anaerobic-aerobic chlorinated ethene degradation field and laboratory studies were carried out at three remediation different sites (Edwards and Cox, 1997). In all three waste sites methane generated from associated waste was proven to be stimulating cometabolism of TCE. The aerobic and anaerobic processes that were impacting the distribution and concentration of contaminants at these sites were confirmed by microcosm studies.
Indigenous sources of carbon associated with soils, sediments and groundwater may be supportive of TCE degradation although certain soils may not support such activities (Fan and Scow, 1993). Walton and Anderson (1990) demonstrated that TCE degradation was carried out by indigenous microbial populations in the absence of an added cosubstrate in rhizosphere and nonrhizosphere soil slurries and groundwater samples from a TCE contaminated site. Although significant differences existed in the rates of TCE cometabolism among methanotrophic microbial communities in these three habitats, similarities existed among the behavior of pure cultures and the indigenous soil microbial populations that cometabolize TCE. Questions still remain as to the specific populations of microorganisms that are responsible for aerobic and anaerobic degradation of TCE in various habitats.
The growth of vegetation encourages the proliferation of microorganisms in the root zone by providing an environment conducive to microbial growth. The density and diversity of microorganisms in the root zone (rhizosphere) is enhanced as compared to bulk soil. Plants also provide an environment that often leads to increased rates of microbial degradation in the rhizosphere of organic contaminants (Walton and Anderson, 1990). Enhanced biodegradation rates may be related to nutrients present in root exudates that are reflected in the increased numbers of microorganisms and/or the increased microbial activity (metabolic or co-metabolic) caused by root exudates. In some cases the TCE groundwater contamination is close to surface soils or impacts seeplines where plumes of contaminated groundwater can emerge as a result of geological and topographical interactions. Significantly higher numbers of methanotrophic bacteria were observed in rhizosphere soils and on roots of Lespedeza cuneata (a legume) and Pinus taeda (Loblolly Pine). It was previously demonstrated that rhizosphere soils from these two types of plants showed higher rates of 14C-TCE mineralization compared to nonvegetated soils (Brigmon et al, 1999). Future work could be focused on in situ microbial distribution and densities, and the role of specific populations including methanotrophs in response to TCE-contaminated groundwater seepage through the rhizosphere.
V. Ex Situ: Bioreactors
It is evident that significant progress has been made in the application of bioremediation utilizing methanotrophic bacteria. Methanotrophic bacteria have also been used in bioreactors as a means of complete removal of contaminant rather than transferring it from one form to another. Bioremediation with methanotrophic bacteria in bioreactors has been tested with free, immobilized, and attached cells. Studies using methanotrophs for TCE removal have been carried out in bioreactors with bacteria attached to carbon (Niedzielski et al. 1989), diatomaceous earth (Strandberg et al., 1989), glass (Phelps et al., 1991), and ceramic packing material (Brigmon et al., 1995). Both aquatic and air bioreactors utilizing methanotrophic bacteria have been developed to remove TCE.
Since the methane is the only nutrient source to the microorganisms, sufficient quantities must be supplied resulting in high flow rates through the bioreactors. Because of the large gas demand relative to the poor solubility of methane and oxygen, this mass transfer can present a design challenge. In addition, pure oxygen and methane can potentially form explosive mixtures so appropriate handling methods are necessary. The media size or design used should be tailored to the velocity of flow. Bioreactors with methanotrophic bacteria have been successfully maintained with both mixed cultures (Alvarez-Cohen and P.L. McCarty, 1991) as well as pure cultures (Uchiyama et al., 1992).
As with all microorganisms, maintenance of methanotrophs in a bioreactor or culture requires specific nutrient conditions. Growth factors that can influence methanotrophic activity are methane concentrations, copper concentration, nitrogen source (NO3, NH4+), oxygen supply, pH, temperature, and the origin of the culture or inoculum used. While methanotrophs have been isolated in culture conditions containing 50% methane (Wise et al., 2000), optimal conditions for TCE degradation in methanotrophic bioreactors are between 4 and 20 % (Strandberg et al., 1989). Bioreactors are useful for further physiological studies of methanotrophs and have added to our knowledge of TCE biodegradation.
Summary
Microbial communities are highly diverse and capable of conducting an extensive range of metabolic activities (Fliermans and Balkwill, 1989). Irrespective of depth or geological formation, subsurface microorganisms carry out all the major nutrient cycling, i.e., carbon, sulfur, nitrogen, manganese, iron and phosphorus. Although each geological formation appears to have its own microbial structure, sandy formations that are highly permeable to air or water flow have higher microbial activity. Considering a generally large subsurface microbiota there is considerable interest for the prospect of degrading hazardous contaminants in situ by stimulating selective bacterial populations (biostimulation) or by the addition of organisms to contaminated sites (bioaugmentation). Stimulation of an indigenous population of methanotrophs by methane is likely to enrich for species that are well adapted to their environment, whereas the deliberate addition of more microorganisms into such an environment may be compromised since the introduced organisms are not as likely to be able to compete.
Defining of temporal and spatial relationships and population dynamics/interactions of selected microorganisms such as methanotrophs in the natural setting is important for the evaluation of bioremediation potential and its effectiveness. It has become increasingly evident that indigenous microbial systems are able to facilitate the degradation and mineralization of a wealth of compounds that twenty years ago were thought to be biologically recalcitrant. This realization has necessitated technologies whereby defined microbial types can be followed in situ in real time by techniques that are designed to be selective, sensitive and easily applicable to soils, sediments and groundwater.
Methanotrophs are physiologically versatile in their ability to exist in a variety of habitats and live in hostile environments having a wide range of pH, temperature, heavy metal concentrations, oxygen concentrations, barometric pressures, salinity and radiation. Under these diverse conditions a number of methanotrophs have been isolated that facilitate the degradation of TCE and its daughter products.
Evaluation, characterization, and utilization of microbial communities associated with in situ bioremediation of subsurface and groundwater contamination is a technological necessity for environmental restoration and assessment. In the past sixty years both industrial and government nuclear production and waste management facilities have generated a significant quantity of organic wastes. These wastes have found their way into the vadose zones and groundwater resulting in unacceptable environmental impacts. The adaptability and manageability of indigenous microorganisms such as methanotrophs make them ideal for the remediation of hazardous environmental wastes under a diverse range of habitats.
References

Figure 1. Oxidation of methane by the methane
monooxygenase enzyme
and associated cometabolism of trichloroethylene.
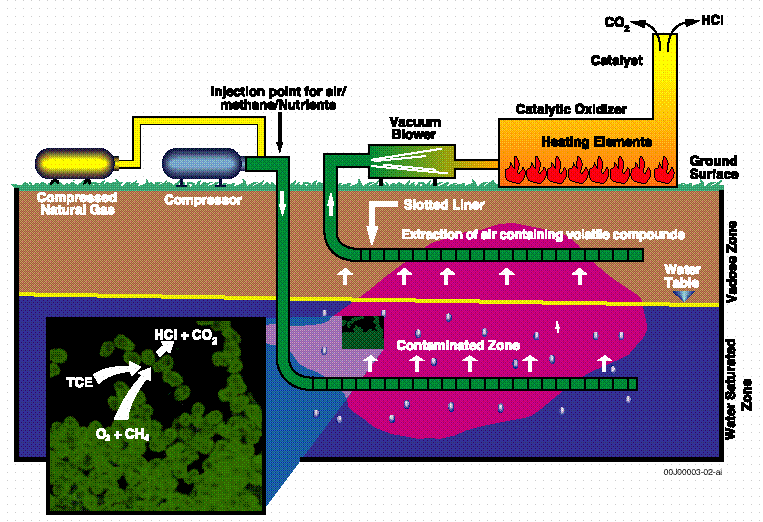
Figure 2. A side view of the horizontal wells
in relation to the surface nutrient injection
and extraction systems (Modified from WSRC, 1992).
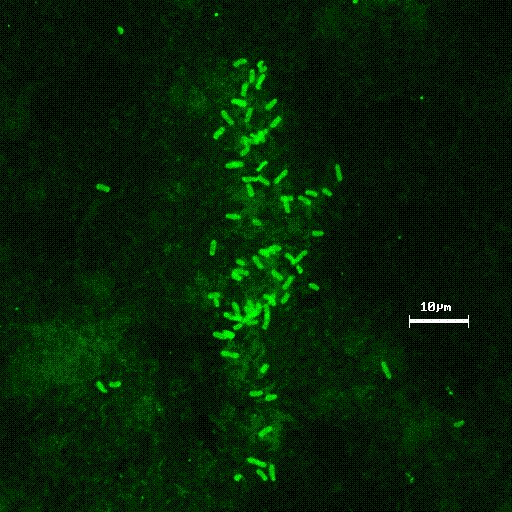
Figure 3. Concentrated groundwater methanotrophic
bacteria on 0.2 m m
filter labeled with fluorescent antibodies.
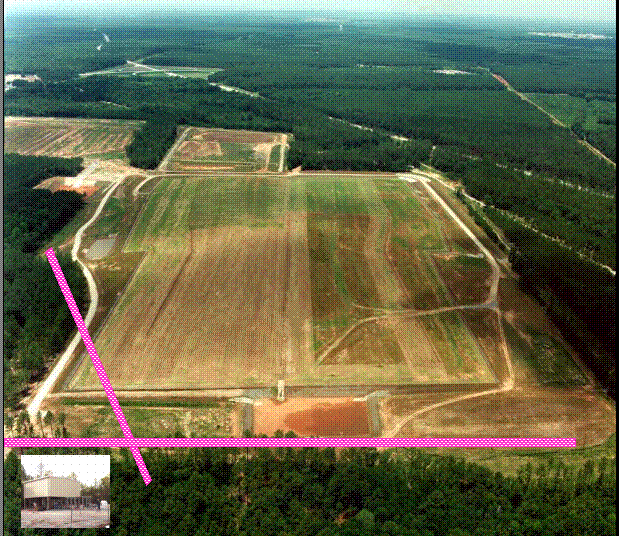
Figure 4. In Situ gaseous nutrient
injection system at the Savannah River Site
70-acre Non-Radioactive Waste Disposal Facility depicting the
400 ft and 600 ft horizontal wells. Insert is
gas-nutrient pumping station.